3dRPC Web Server
3D structure prediction of RNA-protein complexes
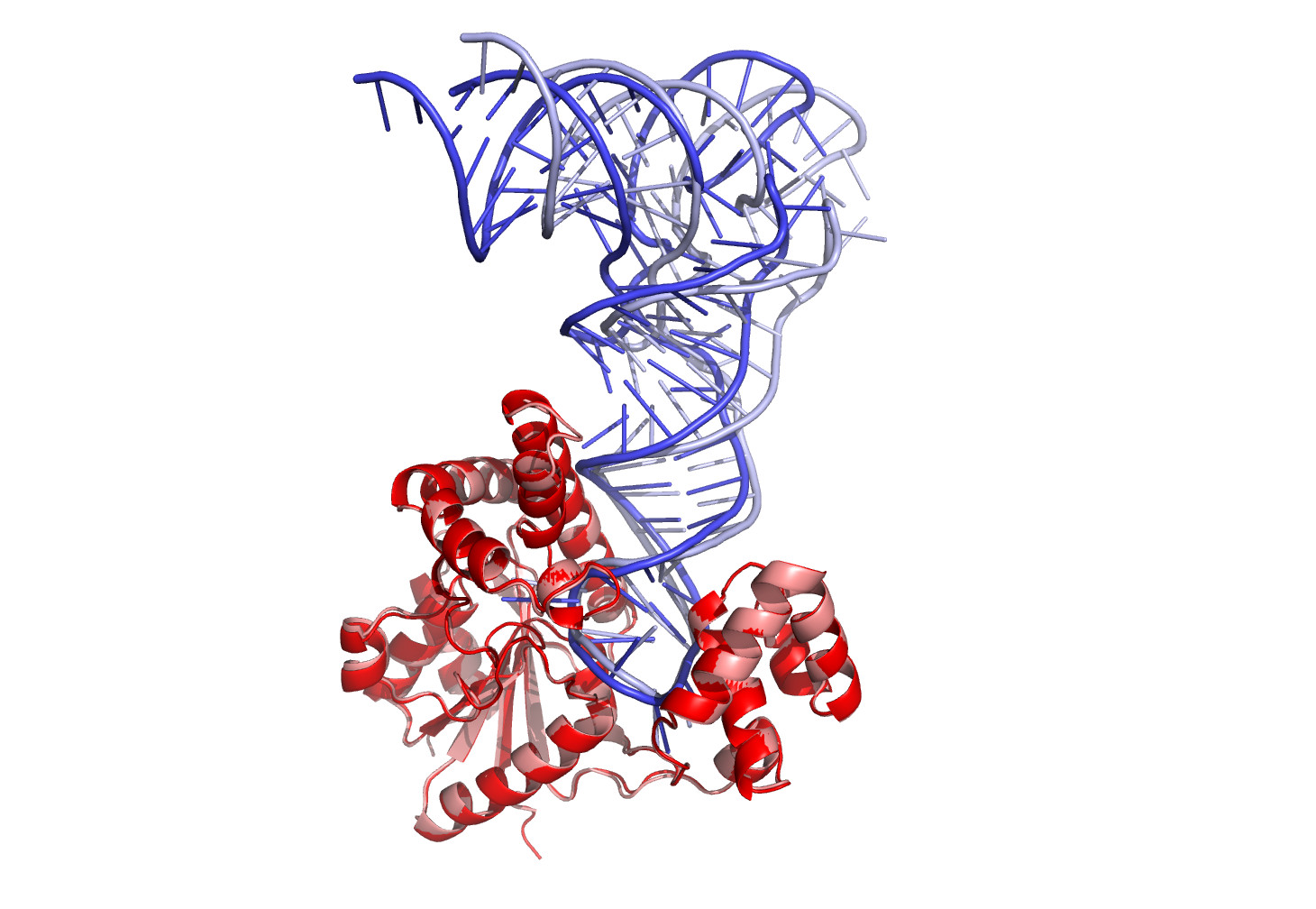
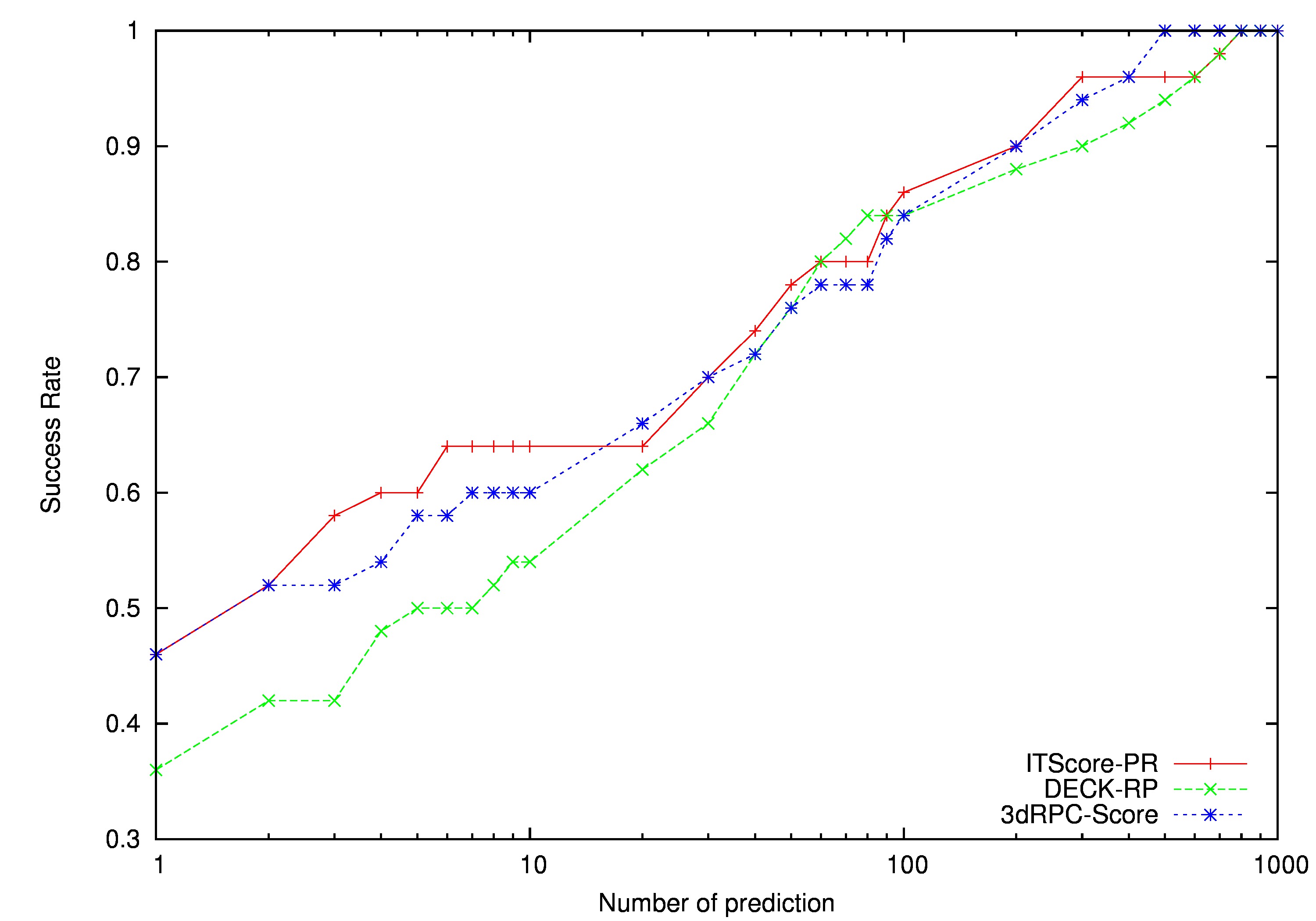
3dRPC is a computational method designed for three-dimensional RNA–protein complex structure prediction. Starting from a protein structure and a RNA structure, 3dRPC first generates presumptive complex structures by RPDOCK and then evaluates the structures by RPRANK. RPDOCK is an FFT-based docking algorithm that takes features of RNA–protein interactions into consideration, and RPRANK is a knowledge-based potential using root mean square deviation as a measure. Here we give a detailed description of the usage of 3dRPC.
You can cite our work like this:
- Huang, Y., et al., a novel protocol for three-dimensional structure prediction of RNA-protein complexes . Scientific Reports, 2013. 3: p. 1887.
- Li, H., Y. Huang, and Y. Xiao, A pair-conformation-dependent scoring function for evaluating 3D RNA-protein complex structures. PLoS One, 2017. 12(3): p. e0174662.
- Yangyu Huang, H.L., Yi Xiao, Using 3dRPC for RNA-protein complex structure prediction. Biophysics Reports, 2016. 2(5-6): p. 95-99.